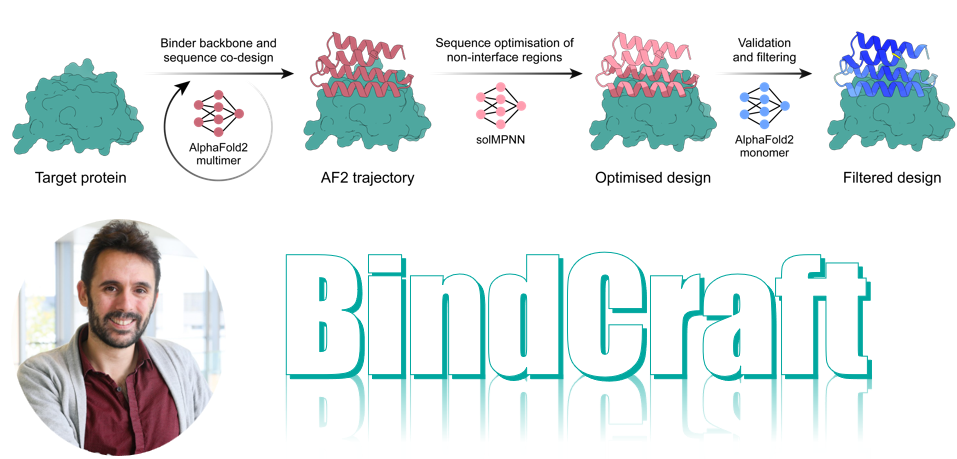
PRODIGY: protein-based molecular design for next-generation therapy
About the project
Proteins are at the heart of every human disease, and are thus targets of most therapeutics, but the development of therapeutic molecules is a difficult challenge.
With the emergence of new machine learning (ML) or generative artificial intelligence (GenAI) methods, enabled by big data, we are at the cusp of one of the most substantial transformations in biomedicine of the last 50 years. Combined with recent breakthroughs in structural biology, in particular cryo-EM, ML-based computational models will enable the design of novel proteins, peptides and small molecule drugs with potentially transformative therapeutic potential for a wide range of applications.
Indeed, GenAI approaches have begun to impact society in various domains such as image and text generation, and are becoming a transformative toolbox in molecular design and drug discovery. Despite these impressive advances, GenAI is however still in its infancy and facing many obstacles, in particular in domains where data is limited, and the problems are complex, such as the design of novel molecular matter acting within a biological context. Many fundamental challenges in ML and GenAI approaches remain unsolved, such as imposing empirical priors, data representation, sampling performance, data availability or explainability, and systematic research efforts are paramount.
Within the NCCR PRODIGY (Protein-based Molecular Design for Next-Generation Therapy), we will set up a collaborative network that combines: excellence in fundamental research in ML methods, computational design of small-molecules, peptides and proteins, structural biology, chemical biology, high-throughput biology powered by omics approaches, to innovate on GenAI approaches for the design of novel therapeutic modalities
Leadership team

Bruno Correia
- EPFL
- Director
- https://www.epfl.ch/labs/lpdi/

Paola Picotti
- ETHZ
- Co-director
- https://imsb.ethz.ch/research/picotti.html

Beat Fierz
- EPFL
- Co-director
- https://www.epfl.ch/labs/lpdi/

Nicolas Thomä
- EPFL
- Deputy co-director
- https://www.epfl.ch/labs/upthomae/

Sereina Riniker
- ETHZ
- Deputy co-director
- https://riniker.ethz.ch/

Matteo Dal Peraro
- ETHZ
- Deputy co-director
- https://www.epfl.ch/labs/lbm/
Our Partners